Microbiological profile of lower respiratory tract infections in neurological intensive care unit of a tertiary care center from Central India
- *Corresponding Author:
- Asst. Prof. Trupti Bajpai
Department of Microbiology, Sri Aurobindo Institute of Medical Sciences Medical College and Post Graduate Institute, Indore, Madhya Pradesh, India.
E-mail: truptiu@rediffmail.com
Abstract
Background: Lower respiratory tract infections (LRTI’s) are the most frequent infections among patients in intensive care units. The consequences of increased drug resistance are far reaching since bacterial infection of the lower respiratory tract (LRT) is a major cause of death from infectious disease. Objective: The study was conducted with the aim of determining the bacterial etiology of LRTI in the neuro intensive care unit (NICU) as well as to update the clinicians with the various antimicrobial alternatives available in the treatment of LRTI. Subjects and Methods: The study was conducted for the period of 3 years from January 2010 to December 2012 in the Microbiology Department of a Teaching Tertiary Care Hospital. The LRT specimens from 230 patients admitted in a NICU during the study period were processed. Following culture, the isolated organisms were identiô€ÂÂÂÂÂÂÂÂÂÂied and antimicrobial sensitivity was performed by standard methods. Results: Out of the 230 LRT specimens evaluated, 198 (86.08%) were culture positive. A total of 254 pathogens were recovered with a predominance of Gram-negative isolates (n = 243; 96.05%) Pseudomonas aeruginosa was the most dominant pathogen followed by Klebsiella pneumoniae. Alarmingly high percentage of extended spectrum beta-lactamase and methicillin resistant Staphylococcus aureus isolates were detected. The resistance to cephalosporins, aminoglycosides and carbapenem were remarkable. Conclusions: Therefore, we can conclude that for effective management of LRTI’s, an ultimate and detailed bacteriological diagnosis and susceptible testing is required to overcome global problem of antibiotic resistance.
Keywords
Extended spectrum beta-lactamase, lower respiratory tract infections, methicillin resistant Staphylococcus aureus
Introduction
Lower respiratory tract infections (LRTI) are the most common bacterial infections among patients in neurological intensive care units (NICUs), occurring in 10-25% of all intensive care unit (ICU) patients and resulting in high overall mortality, which may range from 22% to 71%. Infection and antibiotic resistance are important public health issues. [1-4] One of the major problems world-wide is the increase in antibiotic resistant strains of bacteria, mainly in hospitals and also in the community, which has proved difficult to control without considerable resources and expenditure. [5] The incidence and associated mortality due to LRTI can be influenced by several factors including characteristics of the population at risk, standard of the health-care facilities available, immunosuppressive drugs, inappropriate antibiotic therapy, distribution of causative agents and prevalence of antimicrobial resistance. Highly resistant strains of Gram-negative bacilli (GNB) continue to spread in hospitals causing therapeutic problems in many parts of the world, particularly in developing countries and where isolation facilities for patients with resistant organisms are often inadequate. In developing countries, acute respiratory infection is the leading cause of morbidity and mortality in critically ill patients. [6,7] In almost all cases, eradication of causative agents requires initiation of antimicrobial therapy before obtaining culture report. However, during the last few years, the increase in antibiotic resistance has compromised the selection of empirical treatment. [8] Information on various lower respiratory tract (LRT) bacterial pathogens and their antibiotic resistance patterns in hospitalized patients is inadequate in our country. Hence, the aim of this study was to determine the antimicrobial resistance profile among microorganisms isolated from patients with ICU-acquired respiratory infections, with a futuristic approach toward developing antibiotic policies in association with the clinicians. [7]
Subjects and Methods
The present study was conducted in the Microbiology Department of a Teaching Tertiary Care Hospital during January 2010 to December 2012. The LRT samples (sputum, suction tip, endotracheal, bronchial aspirate and pleural fluid) were obtained from the patients revealing acute respiratory symptoms, admitted in the NICU of the same hospital. The samples were collected aseptically and processed immediately following collection.
Single or mixed growth (two or more than two isolates per specimen) isolated from all the eligible consecutive samples were identified by observing the colony characteristics on the blood agar, MacConkey agar plates and biochemical reactions using standard microbiological methods. [9,10] Isolates from repeat culture of previously recruited patients and isolates identified as commensals or contaminants were excluded. The bacterial isolates were subjected to susceptibility testing by standard Kirby Bauer disc diffusion methods. [11] The susceptibility patterns of the bacterial pathogens were determined following the panel of antimicrobial agents as recommended by Clinical Laboratory Standard Institute (CLSI) -2010. Zone diameter was measured in millimeters and interpreted as per CLSI guidelines. [12] The identification of the bacterial isolates up to species level and their antimicrobial susceptibilities were also confirmed using Vitek 2 compact (BioMérieux, France) The entire testing was done under strict quality control and American Type Culture Collection (ATCC) strains were used as control strains.
The present study has been sent to the ethical committee of our institute for approval.
Results
During the study period, LRT specimens of 230 patients who were admitted to NICU were evaluated. Out of 230 LRT specimens (14 sputum, 136 suction tip, 54 tracheal, 22 bronchial and 4 pleural fluids), 198 (86.08%) were culture positive, whereas 32 (13.91%) specimens showed no growth.
Out of the culture positives, all 198 (100%) specimens showed bacterial isolates while one specimen (0.5%) showed growth of Candida species also.
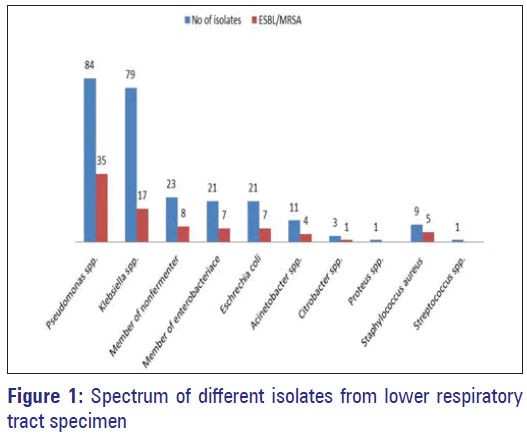
A single pathogen was demonstrated in 150 (75.75%) patients and 48 (24.24%) had mixed bacterial etiology. A total of 254 isolates (including one Candida spp.) were recovered from 198 patients. Out of 253 bacterial pathogens recovered, 243 (96.05%) were Gram-negative and 10 (3.95%) were Gram-positive bacteria. The most prevalent Gram-negative pathogen isolated was Pseudomonas aeruginosa, which was isolated from 84 (33.20%) patients followed by Klebsiella pneumoniae (n = 75 i.e., 31.22%) while the most prevalent Gram-positive pathogen was Staphylococcus aureus (n = 9; 3.55%) followed by Streptococcus spp. (n = 1; 0.39%).
Among 243 Gram-negative bacteria 89 (36.62%) extended spectrum beta-lactamase (ESBL) isolates were recovered while out of nine S. aureus 5 (55.55%) methicillin resistant S. aureus (MRSA) isolates were recovered [Figure 1].
High rates of resistance to cephalosporins (75.38%) were demonstrated by all the Gram-negative bacteria. The susceptibility rates for carbapenem were 75.13% followed by amikacin (71.05%) and Gentamicin (60.45%). The percentage susceptibility of S. aureus and Streptococcus spp. towards vancomycin was 100% and 76%, while that for linezolid was 100% and 72.6% respectively [Table 1].
| Antibiotic | Gram-negative bacilli | |||||||
|---|---|---|---|---|---|---|---|---|
| Organism | ||||||||
| Escherichia coli isolates (21) 07 ESBL | Klebsiella spp. isolates (79)17 ESBL | Pseudomonas spp. isolates (84) 35 ESBL | Proteus Spp (01) bacteriace (21) 07 | Member of Entero ESBL | Acineto bacter (11)04 ESBL | Citrobacter (03) 01 ESBL | Member of non-fermenter (23) 08 ESBL | |
| Amikacin | 49.2 | 68.4 | 63.6 | 100 | 69.2 | 75 | 75 | 68 |
| Ampi/sulb | 47.7 | 53.4 | ND | 00 | 48.1 | 100 | 75.2 | 72.3 |
| Aztreonam | 15.5 | 25.3 | 00 | 00 | 33.3 | 00 | 00 | 30 |
| Cefazolin | 15.5 | 45 | 33.3 | 00 | 00 | 25.2 | 50 | 17.6 |
| Cefepime | 12 | 50 | 33.3 | 00 | 35.9 | 25.2 | 50 | 17.6 |
| Cephotaxime | 45.5 | 50 | 33.3 | 00 | 00 | 00 | 00 | 00 |
| Ceftazidime | 26.9 | 26.5 | 00 | 00 | 33.3 | 50 | 50 | 19 |
| Ceftriaxone | 7.6 | 7.6 | 33.3 | 00 | 38.9 | 00 | 00 | 23.5 |
| Chlorampheni | 54 | 61.3 | 33.3 | 100 | 47.2 | 75 | 75 | 45.2 |
| Ceftizoxime | 00 | 00 | 33.3 | 00 | 33.3 | 20 | 00 | 5.8 |
| Cephoxitin | 38.4 | 38 | 33.3 | 100 | 25.1 | 00 | 00 | 17.6 |
| Cephalothin | 00 | 00 | 33.3 | 100 | 00 | 00 | 00 | 64.7 |
| Gentamicin | 47.7 | 71.2 | 57 | 00 | 67.7 | 75 | 80 | 89.2 |
| Imipenem | 71.7 | 70 | 72.6 | 100 | 83.3 | 100 | 80 | 85 |
| Meropenem | 70 | 76.8 | 68 | 100 | 86.3 | 50.5 | 80 | 83 |
| Netilmicin | 46.6 | 66 | 50.8 | 00 | 44.4 | 35.7 | 00 | 52.9 |
| Tetracycline | 48 | 67.6 | 00 | 00 | 83.5 | 75 | 80.2 | 78.9 |
| Tobramicin | 61 | 63.8 | 48.7 | 00 | 80 | 60 | 75.9 | 80 |
| Trimethoprim | 40 | 50 | 00 | 00 | 00 | 65 | 65.3 | 41.1 |
| Norfloxacin | 30.7 | 32 | 20.8 | 100 | 33.3 | 50.2 | 00 | 41.1 |
| Ticarcillin | 7.6 | 36.3 | ND | ND | 66.6 | 35 | 50 | 56.6 |
| Ampicillin | ND | 51 | ND | ND | 33.3 | 75 | 75 | 47 |
| Piperacillin | ND | ND | 60 | ND | ND | ND | ND | ND |
| Piper/Tazobact | ND | ND | 72.5 | ND | ND | ND | ND | ND |
| Polymixin-B | ND | ND | 91 | ND | ND | ND | ND | ND |
| Ciprofloxacin | 15.3 | 50 | 20.8 | 100 | 55.5 | 50 | 65 | 52.5 |
| Cefmetazole | 23 | 56.8 | ND | 100 | 66.6 | 98.2 | 85.5 | 52.9 |
| AMS | 30.7 | ND | 50 | ND | 22.9 | ND | ND | ND |
| Gram-positive cocci | ||||||||
| Staphylococcus (09) 05 MRSA | Streptococcus spp. (01) | |||||||
| Chlorampheni | 77.7 | 45.2 | ||||||
| Gentamicin | 55.5 | 48.3 | ||||||
| Tetracycline | 55.5 | 46 | ||||||
| Tobramicin | ND | 100 | ||||||
| Clindamicin | 33.3 | ND | ||||||
| Levofloxacin | 55.5 | 58.3 | ||||||
| Ampicillin | ND | 100 | ||||||
| Erythromycin | 33.3 | 56.3 | ||||||
| Rifampin | 55.5 | 43.6 | ||||||
| Vancomycin | 100 | 76 | ||||||
| Moxifloxacin | 42.8 | ND | ||||||
| Teicoplanin | 66.6 | ND | ||||||
| Linezolid | 100 | 72.6 | ||||||
AMS: Antimicrobial stewardship, MRSA: Methicillin resistant Staphylococcus aureus, LRT: Lower respiratory tract, ESBL: Extended spectrum beta-lactamase
Table 1: The percentage antimicrobial susceptibility of the LRT bacterial isolates towards the various antimicrobial agents
Discussion
The main objective of this study was to investigate various isolates from LRTI patients in NICU and to determine the antimicrobial resistance pattern of bacteria against some commonly used antibiotics.
Ventilator associated respiratory infections continue to be a frequent and fatal complication in critically ill patients with mortality ranging from 40% to 80%. [13]
The National Nosocomial Infections Surveillance (NNIS) of the center for disease control of USA reports 60% of nosocomial pneumonias to be caused by aerobic GNB. We found GNB to be the predominant organism (96.04%) with low isolation of S. aureus. These results were similar to those obtained by Veena Kumari et al., Okesola and Ige and Goel et al. who found that GNB isolated was 92.2%, 93% and 97.4% respectively. [1,2,14]
Among GNB, P. aeruginosa (33.20%) was the most common isolate identical to the study made by Goel et al., the results being 35%. Similar results were also quoted by Jarvis and Martone, Gilligan, Jarlier et al. [1,15-18]
It was found that 13.91% of the specimens remained sterile on culture probably due to previous antibiotic therapy or being non-representative specimens. Advances in the medical and surgical manipulations and increase in their applications provide a suitable environment for nosocomial fungal infections. Though in the present study, Candida species was isolated in only one (0.5%) patient, the cases should not be overlooked in future. There was an overall preponderance of GNB among the LRT infection isolates with P.aeruginosa, K.pneumoniae and non-fermenting GNB as the common isolates as also confirmed from the studies made by Veena Kumari et al. (2007). [2]
Pneumonia is a frequent complication in patients admitted to the ICU. It is frequently polymicrobial with predominantly multi-drug resistant GNB such as P.aeruginosa, K.pneumoniae, non-fermenter GNB and Escherichia coli. Incidence of mixed bacterial infection in this study was 24.24% and this is consistent with the fact that the incidence of mixed infections does not usually exceed 30% as has observed in other series (de Roux et al. 2006). [19] However, the identification of polymicrobial infections is very important for treatment strategies and to avoid a false impression of critically resistant strains.
Antibiotic resistance is a major problem in ICU admitted patients. We noticed a high rate of resistance to cephalosporins among the various Gram-negative isolates. Similar observations were made by various reporters including Sofianou et al., Veena Kumari et al. and Goel et al. [1,2,20]
High rate of resistance at our center might be due to the selective influence of extensive usage of third generation cephalosporins. Carbapenems are frequently used as a last choice in treating serious infections caused by GNB’s. Our study showed 24.87% resistance towards carbapenem in accordance to observations made by Akhtar [21] that showed 26.1% resistance and Fatima et al. [15] that showed 24% resistance. This was in contrast to observations made by Gonlugur et al. [22] and Gladstone et al., [23] that comparatively showed lower rates of resistance toward carbapenem, whereas study done by Kucukates and Kocazeybek showed 100% sensitivity to carbapenem. [24] This finding suggests that carbapenem should be judiciously used in ventilated patients to prevent any further increase in resistance.
The resistance of some GNB to aminoglycosides to a longer extent to gentamicin than to amikacin has been well-recognized in many hospitals. In this study, the resistance of Gram negative isolates toward amikacin (28.95%) and gentamicin (39.55%), which is again alarming. The causes behind the emergence of such organisms have been a matter of speculation. Due to high rate of progression of resistance to amikacin, a strategy of limited and prudent use of antibiotics is urgently needed. [4] Aminoglycoside resistant strains are more common at sites with poor penetration of drugs.
S. aureus is known to be a common cause of nosocomial lung infection. In our study, MRSA accounted for 55.55% of nosocomial infections, which was equivalent to NNIS data (52.3%). [25]
No ESBL data for comparison was available as most of the authors did not concentrate on studies related to phenotypic ESBL detection.
However, the rising percentage of ESBL’s (36.62%) and MRSA’s (55.55%) in our study is alarming.
We conclude that multidrug resistant Pseudomonas and Klebsiella are the most common etiological agents of LRTI’s in ICU. There is an alarmingly high rate of resistance to cephalosporins, beta lactam-beta lactamase inhibitors and carbapenem against predominant organism.
The increasing resistance to antibiotics by respiratory pathogens has complicated the use of empirical treatment with traditional agents [26] and a definitive bacteriological diagnosis and susceptibility testing would, therefore, be required for effective management of LRTI. [27]
Now it is well known that critically ill and elderly patients are at greater risk of contracting GNB-LRT infection. Antimicrobial resistance monitoring helps in optimization of antimicrobial therapy and is more important in the ICU’s as infection and antimicrobial consumption are significantly higher. [28]
Acknowledgment
The authors wish to thank the Chairperson and Dean of the institute for providing laboratory facilities and healthy working atmosphere during the study period. The authors are also thankful to the technical staff of the institute for providing necessary helping hand during the endeavor.
References
- Goel N, Chaudhary U, Aggarwal R, Bala K. Antibiotic sensitivity pattern of gram negative bacilli isolated from the lower respiratory tract of ventilated patients in the intensive care unit. Indian J Crit Care Med 2009;13:148-51.
- Veena Kumari HB, Nagarathna S, Chandramuki A. Antimicrobial resistance pattern among aerobic gram-negative bacilli of lower respiratory tract specimens of intensive care unit patients in a neurocentre. Indian J Chest Dis Allied Sci 2007;49:19-22.
- Hospital-acquired pneumonia in adults: Diagnosis, assessment of severity, initial antimicrobial therapy, and preventive strategies. A consensus statement, American Thoracic Society, November 1995. Am J Respir Crit Care Med 1996;153:1711-25.
- Gonlugur U, Bakici MZ, Ozdemir L, Akkurt I, Icagasioglu S, Gultekin F. Retrospective analysis of antibiotic susceptibility patterns of respiratory isolates of Pseudomonas aeruginosa in a Turkish University Hospital. Ann Clin Microbiol Antimicrob 2003;2:5.
- Ayliffe GA, Fraise AP, Geddes AM, Mitchell K. Control of Hospital Infection: A Practical Handbook. 4th ed. London: Arnold; 2000. p. 1-10.
- Navaneeth BV, Belwadi MR. Antibiotic resistance among gram-negative bacteria of lower respiratory tract secretions in hospitalized patients. Indian J Chest Dis Allied Sci 2002;44:173-6.
- Pittet D. Nosocomial pneumonia: Incidence, morbidity and mortality in the intubated-ventilated patient. Schweiz Med Wochenschr 1994;124:227-35.
- Pfaller MA, Korten V, Jones RN, Doern GV. Multicenter evaluation of the antimicrobial activity for seven broad-spectrum beta-lactams in Turkey using the Etest method. Turkish antimicrobial resistance study group. Diagn Microbiol Infect Dis 1999;35:65-73.
- Forbes BA, Sahm DF, Weissfeld AS, editors. Bacterial identification flow charts and schemes: A guide to part III. Bailey and Scott’s Diagnostic Microbiology. 12th ed. Missouri: Mosby Elsevier; 2007. p. 251-3.
- Cheesbrough M. Medical Laboratories Manual for Tropical Countries. Vol. 2. London: Tropical Health Technology, Butterworth; 2002. p. 479.
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966;45:493-6.
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing. 20th Information Supplement. NCCLS document M100-S20. Clinical and Laboratory Standards Institute; 2007.
- Andrew BR, Juan OB. Diagnostic strategies for ventilator associated pneumonia. Contemp Surg 2003;59:178-82.
- Okesola AO, Ige OM. Trends in bacterial pathogens of lower respiratory tract infections. Indian J Chest Dis Allied Sci 2008;50:269-72.
- Fatima A, Naqvi SB, Khaliq SA, Perveen S, Jabeen S. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infections. Springerplus 2012;1:70.
- Jarvis WR, Martone WJ. Predominant pathogens in hospital infections. J Antimicrob Chemother 1992;29 Suppl A: 19-24.
- Gilligan PH. Pseudomonas and burkholderia. In: Murray RR, Baron EJ, Pfaler MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. Washington, DC: American Society for Microbiology; 1995. p. 509-19.
- Jarlier V, Fosse T, Philippon A. Antibiotic susceptibility in aerobic gram-negative bacilli isolated in intensive care units in 39 French Teaching Hospitals (ICU study). Intensive Care Med 1996;22:1057-65.
- de Roux A, Ewig S, García E, Marcos MA, Mensa J, Lode H, et al. Mixed community-acquired pneumonia in hospitalised patients. Eur Respir J 2006;27:795-800.
- Sofianou DC, Constandinidis TC, Yannacou M, Anastasiou H, Sofianos E. Analysis of risk factors for ventilator-associated pneumonia in a multidisciplinary intensive care unit. Eur J Clin Microbiol Infect Dis 2000;19:460-3.
- Akhtar N. Hospital acquired infections in a medical intensive care unit. J Coll Physicians Surg Pak 2010;20:386-90.
- Gonlugur U, Bakici MZ, Akkurt I, Efeoglu T. Antibiotic susceptibility patterns among respiratory isolates of gram-negative bacilli in a Turkish University Hospital. BMC Microbiol 2004;4:32.
- Gladstone P, Rajendran P, Brahmadathan KN. Incidence of carbapenem resistant nonfermenting gram negative bacilli from patients with respiratory infections in the intensive care units. Indian J Med Microbiol 2005;23:189-91.
- Küçükates E, Kocazeybek B. High resistance rate against 15 different antibiotics in aerobic gram-negative bacteria isolates of cardiology intensive care unit patients. Indian J Med Microbiol 2002;20:208-10.
- National nosocomial infections surveillance (NNIS) system report, data summary from January 1992-April 2000, issued June 2000. Am J Infect Control 2000;28:429-48.
- Guthrie R. Community-acquired lower respiratory tract infections Etiology and treatment. Chest 2001;120:2021-34.
- Anderson H, Esmail A, Hollowell J, Littlejohns P, Strachen D. Epidemiologically based needs assessment: Lower respiratory disease. DHA Project Research Programme. Government Publication Publisher: NHS Management Executive; 1993. p. 6-12.
- Leonid SS, Galena RK, Olga SU, Elena CP. Antimicrobial resistance patterns among gram-negative bacilli isolated from patients in intensive care units: Results in multicentre study in Russia. Clin Microbiol Infect 1998;9:497-507.